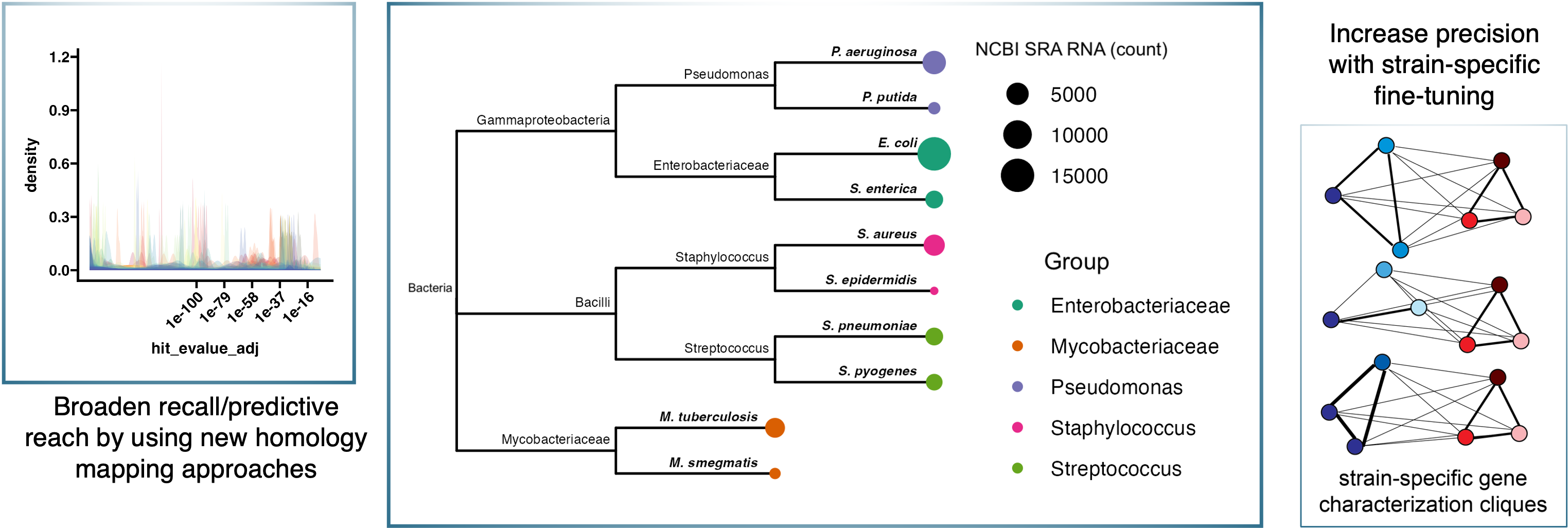
Phylogenetically-guided Transfer Learning
We are exploring the utility of using large compendia of gene expression data from well-studied microbes to boost the performance of machine learning models trained to study microbes with fewer publicly available gene expression datasets. Several compendia of gene expression data have been constructed for model microbes including E. coli, P. aeruginosa, S. aureus and S. cerevisiae (reviewed by the Greene Lab in Lee et al 2023 ). For each of these species there are also gene expression datasets for less often profiled, but equally intriguing, phylogenetically related species. Transfer learning-based neural network architectures are promising approaches for bridging these related, but hetergenous, datasets.
Our aim is to determine the maximum phylogenetic distance between organisms used in pre-training and fine-tuning datasets that promotes the detection of biologically-driven gene expression patterns. This is an important benchmark in expanding the usefullness of machine learning models in the basic biological investigations of the diversity of microbes that interact with human lives and environments, including the skin commensals investiagted by the Oh Lab.
Browse our zotero group library for relevent background reading.